Species Delimitation Challenges
“Species delimitation” is a computational approach to identifying species units in nature. Identification of these units is critical to many areas in evolutionary biology — systematics, phylogeography, biogeography, ecology, conservation, etc. — as well as having impacts in a broader range of areas, such as human health and epidemiology, natural resource management, and so on. Traditional approaches to species delimitation typically rely on models that identify structure in genomic data and identify “species” in nature by relating this structure to species boundaries.
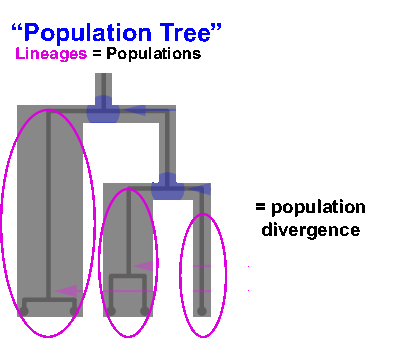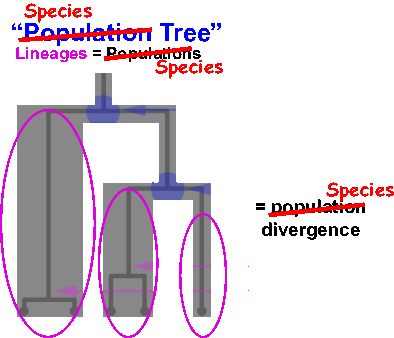
However, other processes may also contribute to the generation of this structure apart from speciation, including population isolation or divergence. This results in “population” units being conflated with “species” units when these approaches are applied without corroborating data. You can learn more about this by watching this Evolution 2017 conference presentation.
This confusion between populations and species has become more and more problematical recently, ironically due to the increasing resolution and scope of the data. As our data increase, our capability to detect finer and finer grained structure below the species level increases, which in turn increases the rate of inflation of artifactual pseudo-species. Various ad hoc, post hoc, or heuristic methods have been proposed to accommodate this within the framework of traditional approach, but they are frankly unsatisfactory in terms of dealing with the actual issue and have their own limits.
A New Paradigm of Species Delimitation Models
Our work pioneers a new age of species delimitation approaches that deal with the problem directly by modeling the issue instead of ignoring it! That is, by actually incorporating an explicit model of the speciation process — in particular, an extended or protracted speciation process — into species delimitation, we are able to discriminate between species and population (or other) boundaries in genomic data. We are just at the beginning stages of this. Our recently submitted paper provides the basic framework for the probabilistic calculations and shows that our approach does indeed work. We want to take this method further, pushing the envelope in several directions, both statistically as well as computationally. To this end, I am looking for students to join me in taking this work to the next level.
Join the Next Generation
Specifically, I am looking for following students:
- A PhD student who is interested in learning Bayesian statistics, MCMC methods, and probablistic modeling to extend our species delimitation model and help implement a Bayesian species delimitation program. The student will receive a stipend, benefits, and coverage of tuition.
If you are interested in being part of this “next generation” revolution of species delimitation appraoaches, please contact me at:
sending me your CV and a description of your background as well as your research interests. You probably should have a look at this page to gain an understanding of what sort of skills you will learn in my lab as well as the work you will do.